News feed
Bayesian direct multimorbidity mapping method using UK Biobank data and artificial intelligence
2017. 09. 06.The first multimorbidity map to indicate comorbidities of 250 most common illnesses has been drawn in Hungary; BME’s VIK involved 120,000 patients to create map.
„We’ve been dealing with methods related to artificial intelligence, machine learning, bioinformatics and biostatistics for over 20 years now. One of those key methods we use is the Bayesian systematic approach that we now applied to map out the degree of indirect mediated comorbidities among various physical disorders”, Péter Antal, associate professor of the Department of Measurement and Information Systems said. He is one of the authors of the article published in PLOS Computational Biology which described the research carried out in collaboration of the Semmelweis University (Budapest, Hungary), Budapest University of Technology and Economics (Budapest, Hungary) and The University of Manchester (Manchester, UK).

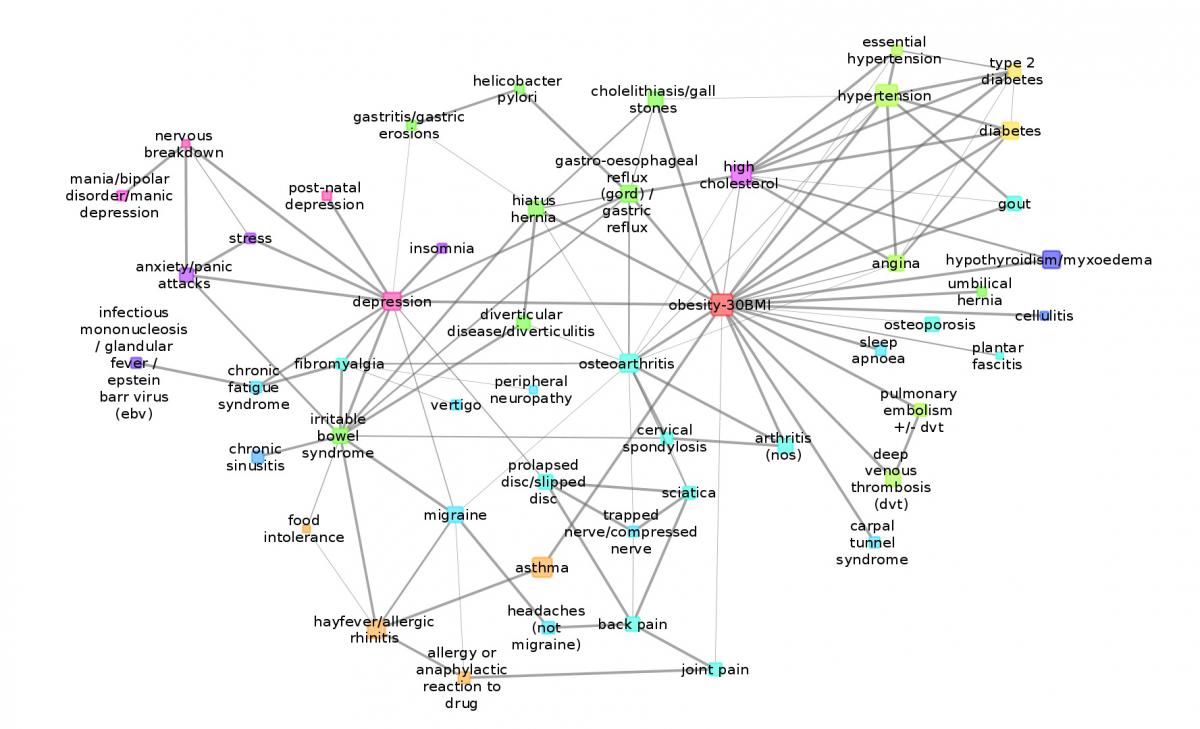
As a result of the work, an electronic map of multimorbodity (a map showing the relationships between various diseases and the impact they exert on one another) has been prepared, demonstrating the 250 most common illnesses with direct mediations. The map can be equally beneficial for physicians, pharmacists and laymen.
The research was based on health information collected from the respondents and stored at UK’s Biobank. UK Biobank is currently hailed as the world’s largest database comprising statistical data of over 500 million volunteers including their genetic profile, previous diseases, their eating and medicine-taking habits, and even some disease-specific data. The research team won access to this database with the help with a previous application.
“The participants provided data to Biobank on a voluntary basis, and their data are strictly protected. There are many other research teams working with our data, but Biobank is careful not to reveal information beyond the professional scope of the applicants, and it only allows them to investigate issues that had previously been requested for” Péter Antal said.
We analysed data collected from 120,000 people suffering from depression. “Our objective was to highlight the different pathways that can lead to depression. We built a model that can indicate what impact one condition – such as depression, obesity, stress or migraine – can exert on others” – continued Péter Antal.
We used a novel statistical method that filters out the 250 most common physical illnesses that was feasible for systematic analysis. PhD student Péter Marx had a key role in the research conducted by the team that the other author of the paper, Péter Antal was also a member of. It was a real challenge to narrow down the diseases as we got all the data in an encoded format. Obesity, for instance, needed to be incorporated into the system taking the subjects’ body/weight index into account, because this condition was not registered as a separate illness in the databank.

Once all the required information was identified, several analyses were conducted simultaneously. One of Péter Marx’s unusual tasks was to transplant the UK codes into the international codes system. To do this, they needed assistance from the field of medical sciences, which they received from co-authors of the paper, György Bagdy and Gabriella Juhász of Semmelweis University.
“To filter the direct impacts of comorbid conditions we used probabilistic graphical modelling, developed at our department” Marx said. This constituted the lion’s share of the work: we had to run computer calculations for several days, which needed to be compared and analysed. We categorised the 250 diseases into 18 groups and received 320 statistically confirmed relationships that did not only mark the link between comorbidity but designated a group of four or five diseases that is worth investigating together.
The paper also reveals the link to the free browsing engine that was in part made up by Péter Marx (link: bioinformatics.mit.bme.hu/UKBNetworks) and which is hosted by the servers at BME’s Department of Measurement and Information Systems. You can search the map of diseases on the interactive platform by specifying variables (i.e. pairs or groups of diseases). If we combine and filter the variables, we can learn whether there is a direct comorbidity between the various illnesses.
The depression map neatly illustrates that this condition is closely linked to several psychiatric disorders but it is also related to obesity and other bodily disorders. Depression, however, is not directly linked to diabetes; its high occurrence with diabetes patients can be linked to obesity.

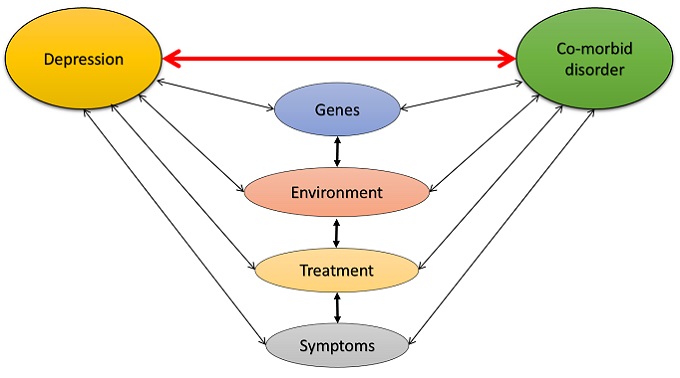
The strength of the relationship between comorbid relations is indicated with lines: the thicker a line is, the stronger the relationship between the specific illnesses. “We still have a few limitations though, – Péter Antal said – as the lines do not indicate (with colours, for instance) whether a relationship is a positive or negative one. One of the most striking findings of recent years was the discovery of inverse comorbidity, i.e. a lower-than-expected probability of a disease occurring in individuals diagnosed with other medical conditions. This is one of the fields we would like to study in the near future, but from now on we will focus on a new challenge in our project: from May 2018 we will have access to all the genetic data stored at UK Biobank; we will then we have the opportunity to download the genotype data of 500,000 participants – a total of 10 TB – and will be able to conduct systematic research into illnesses, consumption of medicine, environmental effects and lifestyles and their impact on one another, which will really be a giant leap forward in bioinformatics.”
HA - GI
Photo: János Philip
Illustration: Peter Antal
